Proceedings of the Seventh
International Conference on Information Visualization: IV’03 (London, July), IEEE Press, pp.252-257
Adapting Single-User Visualization Software for
Collaborative Use PDF
Francis T.
Marchese, Jude Mercado, and Yi Pan
Department of Computer Science
Pace University
New York, NY 10038
fmarchese@pace.edu
Abstract
This paper presents our experiences
with adapting single-user visualization software for web-based collaboration. Sun’s
Java JXTA API was used to adapt an open-source molecular visualization program
called Jmol. It was found that by focusing on the program’s graphical user
interface the software could be quickly transformed into a peer-to-peer application.
Our positive experience implies that many useful single-user programs should be
transformable into tools that make collaboration across the web easier to
initiate, more spontaneous, and supported by a wide range of visualization
software.
1.
Introduction
The Internet has made it
possible for individuals or groups to collaborate at a distance. Collaborative
virtual environments (CVEs) have been developed to support a wide range of collaborative
activities including gaming, immersive virtual reality systems for military training,
scientific visualization, and engineering design [1]. Collaborative
visualization software systems include VisAD [2], COVISA [3], Sieve [4], and
others [5-7]. In general, these programs have been developed to provide a
generic set of collaborative tools for data sharing, representation, and
visualization. However, many domain-specific applications do not easily adapt
to a more general class of visualization programs. Yet there are many
single-user programs that not only would be useful collaborative tools but also
could be adapted for collaborative use. Finally, many collaborative systems
require dedicated hardware and software, even though many collaborations may be
infrequent, of short duration, take place at odd hours, or employ an amalgam of
mainstream conferencing and problem-specific software products.
Therefore, we decided to
investigate these issues by adapting a domain-specific, single-user program for
collaborative use. The goal was to create web-based software that did not rely
on special hardware and could be used at any time, in any place. We selected an
open-source molecular visualization program written in Java called Jmol [8] and
Sun’s Java peer-to-peer (P2P) toolkit – JXTA [9 - 11]. When the project began,
we had little experience with peer-to-peer systems and had never seen Jmol’s
source code. Some of our experiences are presented herein.
The following section
contains the background for the project. Section 3 contains a discussion of
peer-to-peer computing with JXTA. Section 4 presents the process of adapting
Jmol for collaborative use, and Section 5 a sample session for running Jmol in
peer-to-peer collaboration. Discussion and conclusions are found in Section 6.
Section 7 makes suggestions for future work.
2.
Background
Molecular visualization is a
traditional example of a domain-specific visualization application. Molecular
visualization environments tightly integrate data generation, visualization,
and analysis. Moreover, they rely on graphical representations not found in
main stream visualization systems. The
process of displaying electron densities, molecular vibrations, and protein
structures has evolved since the early days of computer graphics in the
mid-1960s. Today, individual or small clusters of single-user, PC-based molecular
visualization systems can compute and render molecular structure and dynamics
in near real-time.
A number of collaborative
molecular visualization systems have been built, including PaulingWorld [12],
Chimera [13], and Mice [14]. PaulingWorld is a virtual reality application designed
to work on SGI workstations with immersive hardware systems such as CAVEs,
Collaborative Workbenches, and head mounted displays. Alternatively, MICE
(Molecular Interactive Collaborative Environment) is a web-based, client-server
system built on Java3D and CORBA. Users translate molecular structure data into
VRML geometry files and load them onto the server. These files are then
collaboratively viewed through Java clients. Finally, Chimera is a
client-server collaborative system that wraps a collection of legacy molecular
analysis and visualization programs within a GUI.
These systems illustrate a
number of issues related to collaborative systems. First, many CVEs require
special purpose hardware and software. This is the case with PaulingWorld,
which runs only on SGI computers attached to VR hardware. Second, CVEs may
require non-native data models. For example, Mice transforms molecular
structure into VRML objects comprised of geometric primitives such as polygon
meshes and surface patches. This loss of original geometry means there is no
way to quantitatively interrogate the source data; thus leaving collaborators
with only the ability to rotate, translate, or scale objects. Finally, although
many CVEs allow users to directly manipulate source data, they are
client-server applications. Chimera allows users access to the original
molecular data, but like Mice, it is a client-server application, and as such,
its collaborative use is subject to constraints. Since the server is
responsible for data transmission, it must have a resolvable IP address for
clients to make a connection. The Internet’s growth has left fewer machines
with static IP addresses, resulting in powerful computing systems assigned
dynamic IP addresses that may inhibit their abilities to be used as servers.
And client-server applications cannot traverse firewalls. Firewall technology
is designed to protect private information. However, there is a need for users
to communicate through a firewall in order to perform collaborative research.
Hence, collaborative
molecular visualization software should support direct access to source data
and metadata, manipulation of native visual representations, interactive data
query and update, and anytime-anywhere collaboration. One possible way to
create such an application is to begin with a pre-existing single user
visualization system and adapt it for P2P use. Because molecular scientists typically
recreate their software with each new generation of technology, there are many
molecular visualization applets written in Java, used to embed molecular
visualizations within web pages [8, 15-16]. One such program is Jmol [8].
JMol is an open-source
molecular visualization program. Chemical scientists use Jmol as a visualization
and measurement tool. It is capable of animating the results of simulations and
can perform transformations (rotation, translation, scaling) upon a molecular
structure. In addition, the application can also measure inter-atomic distances, bond angles, and dihedral angles,
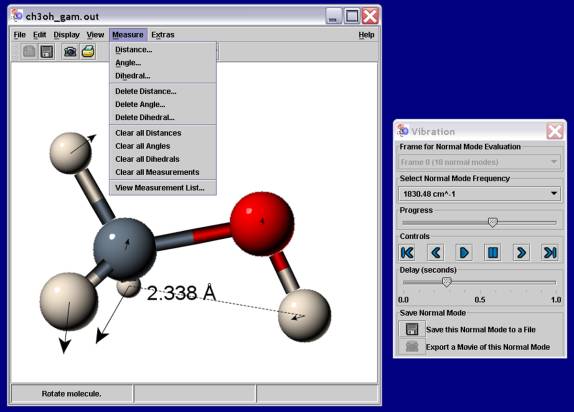
as well as print and export captured images. A snapshot of a Jmol session is
displayed in Figure 1.

The advantage of
using a Java-based program is that it can be paired with JXTA, Sun’s Java peer-to-peer protocol that allows
any network-connected device to communicate and collaborate. Peer-to-peer
networking solves client-server problems by interconnecting machines so that
each node acts as both server and client. There is no single center in a P2P network
as there is in a client-server network. P2P services are distributed, promoting
the robustness of the entire network by eliminating the static IP address
requirement for data transmission. In
addition, with multiple protocols to select for P2P applications and certain
types of peers in use, P2P application users can communicate through a
firewall.
3.
JXTA Background
JXTA is a P2P networking framework, which consists of
three layers: Platform, Services, and Applications. Plaform is the core layer
with the elements for every P2P solution. Services provide the access to JXTA
protocols. Applications use services to access the JXTA network and utilities.
JXTA’s goal is to support P2P programming on any
device from a PDA to a toaster regardless of where it may be located.
Conceptually, it is designed to organize peers and provide an infrastructure
for communication. JXTA does this with protocols expressed in XML.
JXTA is founded on the following basic concepts:
peer, peer group, endpoint, pipe, network transport, advertisements, protocols,
and discoveries.
A peer is a virtual
communication point. There are three basic types of peers: simple peer,
rendezvous peer, and router peer. A simple peer has the least functionality,
and is used behind a firewall. A rendezvous peer processes queries from other
peers, and is used when content transmission is required. A router peer enables
peers to communicate with other peers separated by a firewall.
A peer group organizes peers, and provides specific
services to group members. Data may be shared within the group’s scope, and
peers may check another peer’s status before it is allowed to join the group,
subject to security requirements.
Network transport manages
data transmission over the network. JXTA allows peers to choose different
protocols to fit specific needs. HTTP protocols are used for peers to
communicate through firewalls, while TCP is chosen for intranets.
A network transport system
is composed of endpoints, pipes, and messages. An endpoint refers to an address
of a peer. A pipe is a unidirectional, asynchronous, and virtual connection of
two or more endpoints. Messages contain the data being transmitted. Transmitted
data is packed into a message, which is then sent over the output pipe. At the
other end of the pipe, a peer receives the message from its input pipe and
extracts the transmitted data.
An advertisement is a
structured representation of an entity, service, or resource made available by
a peer or peer group as a part of a P2P network. It is an XML document in JXTA,
containing the description of a message, peer, peer group, or service. Peers
discover advertisements on the network to find other peers. They can use a
cached advertisement, rendezvous peer, or router peer to discover each other
within a LAN or through a firewall.
JXTA has a number of protocols for
advertising, sending, routing, propagating, and securing messages. These
protocols are used to join a peer group, find another peer, create pipes
between peers, and propagate messages among peers. These protocols are used to
effect peer-to-peer collaboration.
A typical scenario for P2P
collaboration using JXTA is as follows:
1.
Start
JXTA
2.
Create
and publish pipe advertisements
3.
Locate
and use the created pipe advertisement
4.
Send
outgoing and listen for incoming messages.
An
application initiates collaboration by joining the Net peer group. The default
Net group gives access to the basic JXTA services. When the JXTA platform is
started, it searches for certain configuration files, most importantly the
PlatformConfig file. PlatformConfig is an XML file that records network settings
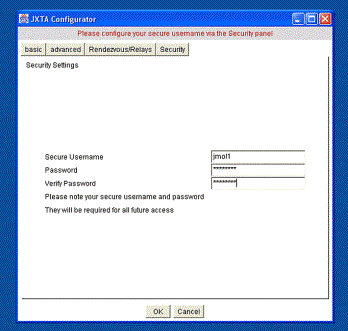
specific to the peer. If this file is not present a JXTA Configuration screen
is shown (Figure 2). It is here that the username, password and advanced
network specifications can be configured.
A Pipe Advertisement is created
using the AdvertisementFactory class. The Pipe Advertisement is an XML file
that contains the type of pipe, the Pipe ID, and possibly an optional name for
the pipe. Once created, peers bind to the pipe’s input and output endpoints to
create instances of the InputPipe and OutputPipe class.
4.
Implementation

Jmol is an event-driven system with
its graphical user interface (GUI) controlling most events. Jmol’s GUI is based
on Java’s Swing interface toolkit (JDK 1.2 and later), a refined version of
Java’s Abstract Window Toolkit (AWT) found in Java’s JDK 1.1 and later. Each
interface component, or widget, is linked to a listener interface that captures
a unique event. Thus, when the mouse is moved, or a button is pushed, or a
dropdown menu item is selected, an event occurs that is captured and acted
upon. And Java’s listener interface provides specific methods in which to embed
actions to be taken once an event has been captured.
By finding the Java
methods in Jmol that control the listener interface it should be possible to
insert the JXTA code necessary
to transmit an event and its attributes to a peer.
Jmol’s interface events result in loading and saving files, updating the
molecular graphic representation, and generating geometric data to describe
inter-atomic distances and angles. After the first peer acts on the event by
updating its local state, it should send a unique message to the second peer
for it to replicate the process. For example, the sending peer captures combined
button and mouse events for translating the molecular representation, and
redraws it at a new screen location. After translation, a message is sent to
the receiving peer, corresponding to a translation action. The receiving peer
then updates its state to match the sending peer.
In addition to updating
Jmol’s event handling methods, three other issues needed to be resolved in order
to transform Jmol into a collaborative system. First, a database model needed
to be selected. A distributed database model was assumed in which each site
retains a copy of all files used in Jmol. The advantage is that file
replication reduces transmission times and creates minimal communication time
lag, since only changes in the program’s state need be transmitted. The logic
behind this is that most molecular data can be easily downloaded or
transmitted, and collaborative users would already have these on their
machines.
Second, JXTA code must
be added to implement the essential JXTA protocols. In particular, communication channels must be
established, incoming messages handled, pipe advertisements discovered, and
output pipes created.
Finally, messages between peers
should be compact in order to minimize communication overhead. For Jmol,
messages only need to include a key specifying an action and an element
containing the attributes of that action. For example, a translation operation
would send the key “XLATE” along with an element containing the translation
matrix. The receiving peer, upon recognizing the message, would use the
transformation matrix to perform a translation locally.
With strategy in hand,
Jmol was adapted for collaboration by adding code to set up the pipes necessary for
communication channels among peers and modifying the graphical user interface
(GUI) components. Upon analysis, it was found that out of the 270 class files
that constitute Jmol, the GUI components are isolated in five files:
DisplayPanel.java, Animate.java, Vibrate.java, and Measure.java.
DisplayPanel.java
contains controls for geometric transformations as well as rendering options. There
are action methods for picking, rotation, zooming, translation, shading, and
the like. Every “Action” method found within DisplayPanel was modified to send
a message to a listening peer for it to perform the same task.
Animate.java
creates the Animate class, which controls the animation of reaction
simulations. It displays VCR controls
that allow the user to start, stop, rewind, forward, jump to a frame, or speed
up the simulation. When a user chooses to ‘Play,’ ‘Rewind,’ ‘Fast Forward,’
etc., a message corresponding to the action is sent to the listening peer so
that it can change the frame on its local application.
Vibrate.java
displays molecular vibrations as animations. As with the Animate class, it
contains controls to specify how a sequnce of frames is displayed. The
actionPerformed() method, which controls animation playback, is augmented to
send a message for every control action performed.
Measure.java is responsible for
measuring angles, distances, and dihedrals between atoms. It also maintains a measurement table
containing the various measurements made by the user. It was modified to send messages that correspond to a specific
“Action.” Actions include measuring or deleting a distance, a bond angle, or
dihedral angle.
Jmol.java
is the main Java class. It was modified to establish communication channels by
implementing theJXTA interfaces: PipeMsgListener, DiscoveryListener, and
OutputPipeListener. PipeMsgListener is used for handling incoming messages;
DiscoveryListener is used for discovering pipe advertisements; and
OutputPipeListener is used when an instance of an OutputPipe is created.
Finally,
Jmol was augmented with a chat facility that provided for saving chats and
reloading for further review.
5.
Running Collaborative Jmol
The collaborative version of
Jmol is implemented as a Java application. Once Jmol has been loaded from a
command line prompt, a dialog box appears. For first time execution, the dialog
box sets up user id and security parameters (Figure 2). Subsequent invocations
bring up only a login dialog. Once logged in, a command line message requests
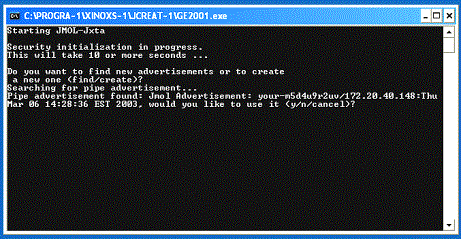
the user to either create an advertisement for a new peer session, or find an
advertisement for an existing peer session (Figure 3). To originate peer
collaboration, the former is selected; otherwise the peer will attempt to find
the originating peer on the network. If the user has selected advertisement,
the Jmol GUI will load and advertise to the netgroup (Figure 4); otherwise Jmol
will search the network looking for a peer connection. When an advertising peer
has been located (Figure 3) the searching peer has the option to make a
connection. Once the connection is made, the Jmol GUI will be displayed.

One peer will load a molecule through the
“File” drop-down menu to begin collaboration. This filename and a “Draw”
message will be sent to the other peer for it to automatically load the
molecule from its local directory. Either collaborator can transform the molecule,
display its properties, or make measurements. For example, when one
collaborator selects rotation, and drags the mouse across the molecular
representation, both see exactly the same view, smoothly rotating in three
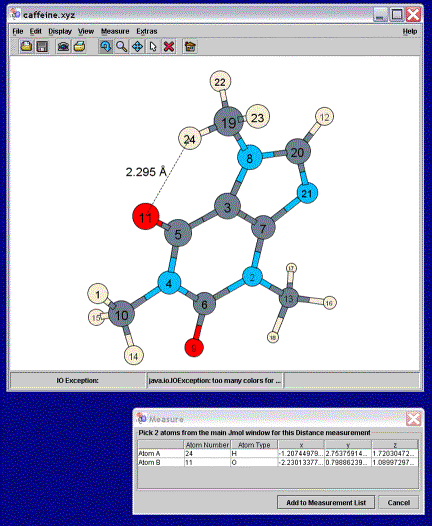
dimensions about the x and y-axes. When a collaborator selects
measure mode, a table appears that is filled with geometric information as the
requisite atoms are selected with the mouse (Figure 5). When the “Add to Measurement
List” button is chosen, the measurement is appended to the image at both
locations. This text is a three-dimensional graphic of geometric information,
and travels along with the molecule as it is rotated, scaled, and translated.


During the course of their engagement,
collaborators may carry on a text chat with the built-in chat facility or use
any other communication tool available for audio or video conferencing, such as
Microsoft’s NetMeeting. When it is time to end the session, all that is
required is to shut down the application.
6.
Discussion and conclusions
The
goal of this project was to create a collaborative visualization application
from a single-user program. Its implementation is consistent with the design
objectives put forth by Wood and coworkers [3]. Collaborative visualization
systems should allow users to share program control, collaborate dynamically,
use the software in an instruction scenario, learn it easily, and exchange data
readily. Collaborative Jmol meets these design goals. Either participant may
control any of Jmol‘s parameters at any time, supporting variability in
visualization scenarios. Jmol allows either participant to work as an instructor,
because each peer may take control of the analysis at any time, and every
action in a sequence is replicated on the collaborating peer. Jmol is easy to
learn because the visualization components of the program remain unchanged;
only menu-driven networking components have been added. Finally, all data is
exchanged between peers, meaning both peers are synchronized in identical collaboration
states.
Peer-two-peer
bi-directional synchronization is executed in Jmol using two asynchronous pipes
by binding each peer to the input end of one pipe and the output end of the
other. When a GUI event occurs, the sending peer transmits a keyword
designating the GUI operation along with its numerical operands through the
input end of the pipe, while the receiving peer extracts the message at the
pipe’s output end. Because these pipes act independently, it is possible for
both peers to transmit messages simultaneously, possibly corrupting visualizations.
Although access to the shared state may be regulated, the current version of
Jmol assumes a free access model similar to cAVS, in which any participant
may affect the common collaborative state at any time [17]. However, since Jmol
will be used as part of a teleconferencing suite, it is expected that such
problems will be mitigated because both participants’ actions will be
coordinated as part of discussion and exploration.
The
process of adapting Jmol was constrained in two ways. First, domain experts,
not software engineers, created Jmol. As such, these individuals typically are
less concerned with a flexible software design that makes for ease of expansion
and adaptation, than with building a product to meet current visualization
needs. Yet, the modularity of Java’s AWT, and its concomitant isolation of
interface methods, made it relatively straightforward to embed the requisite
JXTA code. Second, JXTA was in early development when this research began, today,
version 2 of JXTA is available. Some of its classes were not implemented,
unstable, or undergoing change. Therefore, it constrained the adaptation
process. Despite this, JXTA’s core functionally was sufficiently well developed
to adapt Jmol for basic collaboration.
Jmol
was selected for adaptation because it represented a useful software product
with enough functionality to embody significant program structure and
complexity. This decision was based on experience using the program, not on an
analysis of the source code. It was only after Jmol was selected that its
source code was examined. Then the Jmol transformation was achieved by an
undergraduate computer science major.
What
this research demonstrates is that within a two-month period, working
part-time, an individual with no knowledge of the visualization software, and
learning JXTA on the fly, can transform a single-user program into a functional
and useful collaborative application. Therefore, it follows that a domain
expert, who thoroughly understands the visualization application, could create
a collaborative system within a very short time frame.
In
conclusion, the demonstrated ability of JXTA to quickly create two person,
peer-to-peer collaborative software from single-user visualization systems
means that collaboration across the web will become easier to initiate, more
spontaneous, and be supported by a wide range of visualization software.
7.
Future Work
JXTA
offers the opportunity to have many peers collaborate, but a design change in
Jmol may be required. Presently, there is no visual or auditory cue from Jmol
that reveals which peer is controlling the interface. The natural assumption is
that the other peer is in control. Additional peers create ambiguity. Without
some cue from the software, knowledge of who is in control is uncertain at
best. Even if the collaborators employ audio or video conferencing software as
part of their session, there is no way of knowing with certainty who is
manipulating the model.
This
issue is part of a more general problem with which designers of large-scale
networked collaborative environments must contend – how to provide a shared
sense of presence. CVEs should represent all participants and their actions in
the virtual space they share. In Jmol that would mean at the very least the GUI
should convey who is currently controlling the software. Hence, the next step in
the process of adapting Jmol is to solve this design problem, so the identity
of each user in the collaboration is clearly conveyed.
8.
Acknowledgements
This
project was underwritten in part by National Science Foundation Grant
ANI-0125043. The author would also like to thank Susan Merritt, Dean of the
School of Computer Science and Information Systems, for her continued support.
9.
References
[1]
S.
Singhal and M. Zyda. Networked Virtual Environments. Addison-Wesley,
1999.
[2]
W. Hibbard,
“VisAD: Connecting People to Computations and People to People,” Computer
Graphics 32,3, 1998, pp. 10-12.
[3]
J. Wood, H. Wright, K. Brodlie, “Collaborative
Visualization,” IEEE Visualization '97, 1997, pp. 253-260.
[4]
P. Isenhour, J. Begole, W.S. Heagy, and C.A. Shaffer,
“Sieve: A Java-Based Collaborative Visualization Environment,” IEEE
Visualization '97 Late Breaking Hot Topics Proceedings, October 22-24,
1997, pp. 13-16.
[5]
A. Pang and C.Wittenbrink, “Collaborative Visualization
with CSPRAY,” IEEE CG&A , (March/April) 1997, pp. 32-41.
[6] C. Bajaj and S. Cutchin, “Web based Collaborative Visualization of
Distributed and Parallel Simulations,” Proceedings of IEEE Parallel
Visualization and Graphics Symposium, October 1999, pp. 47-54.
[7]
M.Abbott and L.K. Jain, “DOVE: Distributed Objects
based scientific Visualization Environment,” ACM 1998 Workshop on Java for
High-Performance Network Computing. ACM Press, 1998.
[8]
“Jmol,” http://jmol.sourceforge.net/ (current 23 Mar. 2002).
[9]
“Project
JXTA,” http://wwws.sun.com/software/jxta/ (current 23 Mar. 2003).
[10]
D. Brookshier, D. Govoni, N. Krishnan. JXTA: Java P2P
Programming. Sams, 2002.
[11]
B. Wilson. JXTA. New Rider’s Publishing, 2002.
[12]
S. Su,
R. Loftin, D. Chen, Y. Fang, and Ch. Lin, “Distributed Collaborative Virtual Environment:
PaulingWorld.” Proceedings of 10th International Conference on Artificial
Reality and Telexistence, 2000, pp. 112-117.
[13] C.C. Huang, G.S. Couch, E.F.
Pettersen, and T.E. Ferrin, “Chimera: An Extensible Molecular Modeling Application
Constructed Using Standard Components,” Pacific Symposium on Biocomputing
1, 1996, p. 724.
[14] P.E. Bourne, M. Gribskov, G. Johnson,
J. Moreland, and H. Weissig, “A Prototype Molecular Interactive Collaborative
Environment (MICE),” Pacific Symposium on Biocomputing, Eds. R Altman,
K. Dunker, L. Hunter, and T. Klein, 1998, pp. 118-129.
[15]
“JMV: Java Molecular Viewer,” http://www.ks.uiuc.edu/Research/jmv/
(current 23 Mar. 2003).
[16]
“Java Mage: Kinemages in your web browser,” http://kinemage.biochem.duke.edu/javamage/java.html (current 23 Mar. 2003).
[17]
“Collaborative
AVS,” http://www.tacc.utexas.edu/cavs/overview.html (current 3 May 2003)